Andre Franke
Immunogenetics and Bioinformatics
Mission Statement
The World Health Organization (WHO) ranks chronic diseases as the greatest threat to human health. Chronic inflammation has emerged as the most significant cause of death in the world, and the prevalence of associated diseases is anticipated to increase steadily in the coming years.
Operating at the interfaces between immunology, genetics and bioinformatics, the research group “Immunogenetics and Bioinformatics” studies the complex interplay of intrinsic genetic traits, host microbiome and extrinsic environmental factors that shape the immune system and, when dysregulated, can drive inflammatory diseases, such as inflammatory bowel disease (IBD) and primary sclerosing cholangitis (PSC). Besides patients, we perform also analyses on population-scale cohorts from Schleswig-Holstein and other areas in the world.
T cells are central regulators of adaptive immunity and inflammatory responses. Each T cell expresses a unique T cell receptor (TCR), selected by its ability to bind peptides presented by major histocompatibility complex (MHC) molecules, known as the human leukocyte antigen (HLA) in humans, on the surface of antigen-presenting cells. Structurally, a TCR recognizes both peptide and the MHC protein (pMHC). The TCR-pMHC interaction forms an integral part of the immunological synapse that governs the development, expansion and downstream effector functions of T-cells. Disruption of the balance between the specificity and cross-reactivity among TCR-pMHC interactions can lead to aberrant inflammation and autoimmunity.
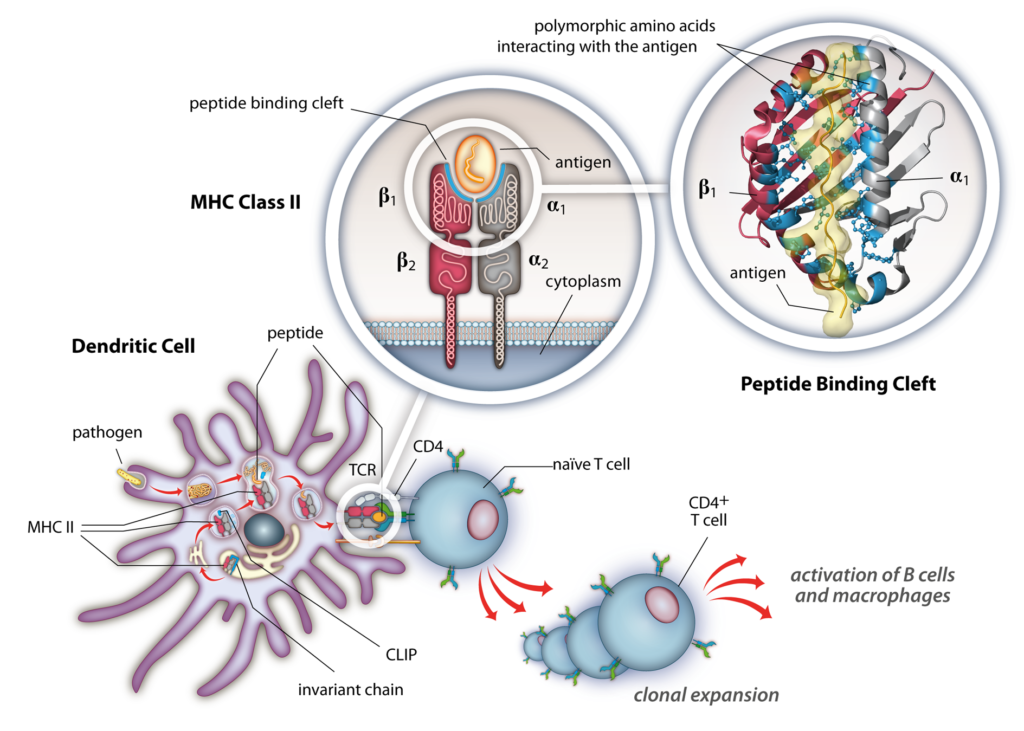
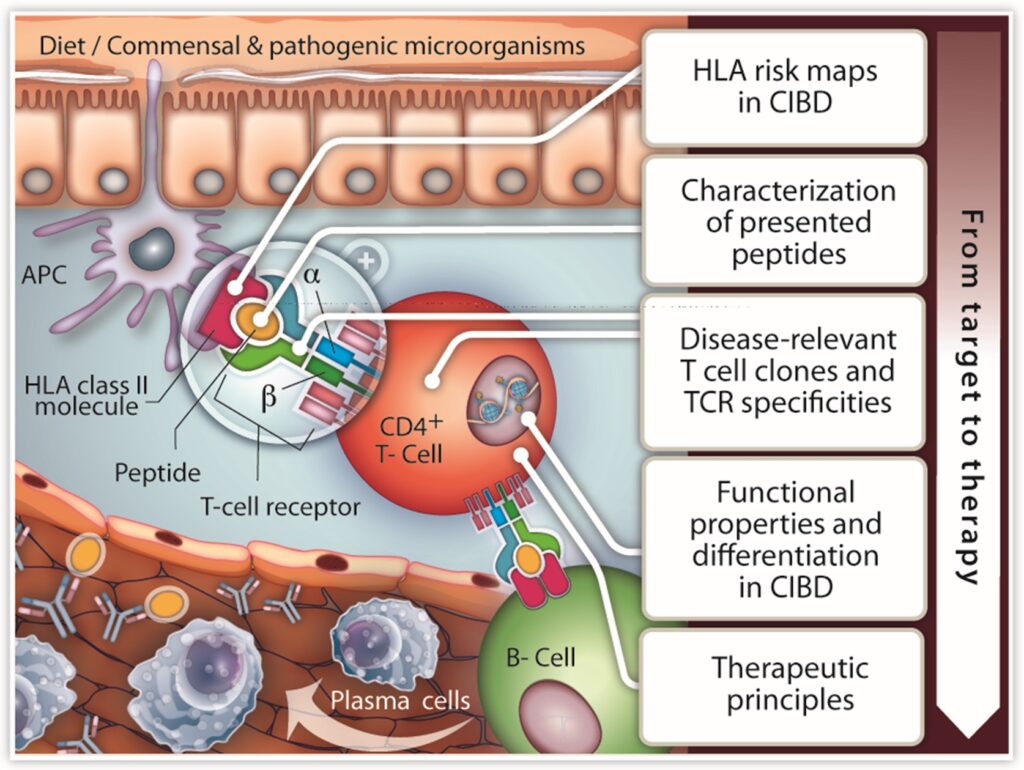
We focus on combining patient-specific HLA risk maps, TCR repertoire profiling and microbiome analyses from large cohorts with antigen discovery technologies to identify pathogenic sources of antigens and to molecularly define actionable targets for disease prevention and treatment.
We employ cutting-edge sequencing technologies and develop HLA-typing bioinformatic tools on large cohorts of patients, healthy individuals and first-degree relatives (FDRs; popgen IBD Family Cohort) to dissect disease-associated signals. We focus on early disease and pre-disease cohorts. In addition, we established bulk and single-cell RNAseq to profile TCR repertoires and characterise specific T cell clonotypes from patient and control material.
To facilitate these studies and to investigate the interaction between the intestinal microbiome and the human host, our Microbiome Core facility uses advanced technologies for biobanking and high-throughput analysis, from sample collection/management to library preparation, sequencing and bioinformatic analysis. Microbe sequencing using 16S Amplicon sequencing and metagenomics is further complimented by culturomics approaches. We envision that our approaches will drive the detection of microbial changes from pre-clinical to disease onset and progression. This, in turn, can help define the functional and causal relationships leading to the identification of therapeutic interventions.
We continue to invest in next-generation technologies for antigen discovery. These involve the development of bioinformatics tools (employing artificial intelligence methods) for the analysis of immunopeptidomic data, the establishment of protocols for antibody profiling such as phage immunoprecipitation sequencing (PhIP-Seq), and antigen-display screens based on yeast and lentiviral platforms to identify peptides recognized by disease-associated T cell receptor (TCR) and MHC molecules. These technologies will drive high throughput identification of novel antigens/peptides associated with autoimmune diseases.
We apply our bioinformatic expertise to layers of data derived from genomics, proteomics, metabolomics, metagenomics, diet, lifestyle and social factors to disentangle the complex pathology of inflammatory diseases. This work forms part of our long-term objectives of a high-resolution and successful application of personalized medicine in inflammation.
Other current research topics of the working group are, for example, data management, eHealth applications (including smartphone apps and wearables for patients; see also the StatusPlus webportal for reporting data back to probands), molecular blood group typing, COVID-19 genetic studies, hemorrhoids disease, and mGWAS (microbiome genome-wide association studies, i.e. mQTL studies) to identify host genetic factors that shape the human microbiome (skin, gut and oral microbiome under study).
We are part of different international consortia, for example, the International IBD Genetics Consortium (IIBDGC), the International PSC Study Group (IPSCSG), nf-core, the COVID-19 Host Genetics Initiative (COVID19 hg), and The Genome Aggregation Database (gnomAD).